Characterization of mycorrhizal fungi of the genus Tulasnella (Tulasnellaceae, Basidiomycota) in the genus of orchids Bletia from Barranca del Cupatitzio Natural Reserve, Mexico
María de los Ángeles Beltrán-Nambo1, Juan Carlos Montero-Castro2, Miguel Martínez-Trujillo3, Rafael Salgado-Garciglia4, J.T. Otero-Ospina5 & Yazmin Carreón-Abud6,*
1,3,6 Laboratory of Genetic and Microbiology, Faculty of Biology, Universidad Michoacana de San Nicolás de Hidalgo, Mexico.
2 Laboratory of Plant Molecular Systematics, Faculty of Biology, Universidad Michoacana de San Nicolás de Hidalgo, Mexico.
4 Laboratory of Plant Biotechnology, Chemical Biological Research Institute, Universidad Michoacana de San Nicolás de Hidalgo, Mexico.
5 Department of Biological Sciences, Faculty of Agricultural Sciences, Universidad Nacional de Colombia, Palmira sede, Colombia.
* Author for correspondence: ycabud@gmail.com, https://orcid.org/0000-0002-6148-6513
1 angelesb2008@gmail.com, https://orcid.org/0000-0003-2240-5933
2 cestrum2003@yahoo.com.mx, https://orcid.org/0000-0002-3098-1415
3 codigogenetico@gmail.com, https://orcid.org/0000-0002-6523-6618
4 rafael.salgadogarciglia@gmail.com, https://orcid.org/0000-0001-5920-6562
5 jtoteroo@unal.edu.co, https://orcid.org/0000-0002-0810-183X
| |
Abstract
Beltrán-Nambo M.A., Montero-Castro J.C., Martínez-Trujillo M., Salgado-Garciglia R., Otero-Ospina J.T. & Carreón-Abud Y. 2018. Characterization of mycorrhizal fungi of the genus Tulasnella (Tulasnellaceae, Basidiomycota) in the genus of orchids Bletia from Barranca del Cupatitzio Natural Reserve, Mexico. Anales del Jardín Botánico de Madrid 75 (2): e075. https://doi.org/10.3989/ajbm.2491
The goal of this study was the identification of mycorrhizal fungi associated with three terrestrial orchids of the genus Bletia Ruiz & Pav.: B. roezlii Rchb. f., B. purpurata A.Rich., and B. punctata Lex., in Barranca del Cupatitzio Natural Reserve—Michoacán, México—. Thirty-nine strains were isolated and morphologically characterized. Nine strains were selected from the molecular analysis. Bletia punctata, an endemic species of Mexico, showed the lowest variability in mycorrhizal fungi. Morphological analysis showed that 39 isolated strains belong to the ‘Rhizoctonia-like fungal complex’. According with the tree of Euclidian distances generated by the analysis WARD, all isolates were included into four subgroups, all related to the genus Tulasnella J.Schröt—which belongs to the ‘Rhizoctonia-like fungal complex’—. Molecular and phylogenetic analysis of the nine selected strains corroborated the results of the morphological study: the sequences obtained were clustered in four subclades related to species of Tulasnella. Our results indicate that a single species of Bletia from a single locality can be associated with different species of mycorrhizal fungi, at least during the adult stage and that the combination of morphological and molecular analyses is a good tool to identify orchid mycorrhizal fungi.
Keywords. ITS, Rhizoctonia-like fungal complex, terrestrial orchids, Tulasnella, orchid-fungal partnership.
|
| |
Resumen
Este estudio tuvo por objetivo la identificación de hongos micorrícicos asociados a tres especies de orquídeas terrestres del género Bletia Ruiz & Pav.: B. roezlii Rchb. f., B. purpurata A.Rich. y B. punctata Lex. en la Reserva Natural Barranca del Cupatitzio —Michoacán, México—. Treinta y nueve cepas fueron aisladas y caracterizadas morfológicamente, de las que nueve se seleccionaron para el análisis molecular. Bletia punctata, endémica de México, presentó la menor variabilidad de morfotipos de hongos micorrícicos. El análisis morfológico demostró que todos los aislamientos pertenecen al ‘complejo Rhizoctonia’. De acuerdo con el árbol de distancias euclídeas generado mediante análisis WARD, todas las cepas se agruparon en cuatro subgrupos, todos relacionados con el género Tulasnella J.Schröt —que pertenece al ‘complejo Rhizoctonia’—. Los análisis molecular y filogenético de las nueve cepas seleccionadas corroboraron los resultados del estudio morfológico: las secuencias obtenidas se distribuyeron en cuatro subclados relacionados con especies de Tulasnella. Nuestros resultados indican que una misma especie de Bletia puede asociarse al mismo tiempo con varias especies de hongos micorrícicos, al menos durante la etapa adulta, y que la combinación de análisis morfológicos y moleculares es una herramienta útil para la identificación de hongos micorrícicos en orquídeas.
Palabras clave. Asociación orquídea-hongo, complejo Rhizoctonia, orquídeas terrestres, ITS, Tulasnella.
|
INTRODUCTIONTOP
The family Orchidaceae Juss. comprises approximately 1,000 genera and 27,000 species around the world (Govaerts & al. 2016). However, a large number of these species is included in some risk extinction category because of direct or indirect human activities (Whitman & Ackerman 2015). The species of this family stablish associations with other organisms during some critical stages of their life cycle, such as pollinators at flowering and symbiotic fungi during germination stages (Rasmussen & Rasmussen 2007; Schatz & al. 2010). Furthermore, it is necessary to considerate that most of the adult orchids have some degree of mycotrophy with mycorrhizal fungi and that the specificity in this kind of association is different between the species of orchids and the partners of fungi , and change according with geographical regions (Rasmussen & Rasmussen 2007; Valadares & al. 2015). In order to implement their reintroduction or programs of sustainable conservation, it is necessary to preserve the essential conditions for the survival of these plants throughout their life cycle.
In American tropical and temperate regions, a substantial number of studies about the diversity of orchid mycorrhizal fungi have been performed for conservation purposes (Valadares & al. 2012, 2015; Otero & al. 2013; Pereira & al. 2014; Nogueira & al. 2014). However, the investigation aimed to mycorrhizal fungi in orchids is uncommon in Mexico (Ortega-Larrocea & Rangel-Villafranco 2007; Ortega-Larrocea 2008; Ortega-Larrocea & González 2008), and little is known about the ecological and phenological aspects, the management for conservation or the reintroduction purposes. Moreover, there are few studies about the identity of the fungal partner or the specificity on endemic orchid species associations in Mexican forests (Ortega-Larrocea & Rangel-Villafranco 2007).
The genus Bletia Ruiz & Pav. includes about 40 species of terrestrial orchids, some of them with a wide distribution range from Mexico to Central America, while others with a more restricted distribution, mostly endemics to Mexico (Sosa 1992). It also includes some species in danger of extinction (Ortega-Larrocea & Rangel-Villafranco 2007), and other that, according with the Official Mexican Norm—NOM-059-ECOL-2010, cf. SEMARNAT (2010)—, could be endangered if adequate actions are not taken. Therefore, this genus can be used to stablish an appropriate methodology for isolation, characterization, and cultivation of fungal partners, and posteriori apply this methodology for conservation of others orchids species at some risk category.
A great number of orchid mycorrhizal fungi has been assigned to the ‘Rhizoctonia-like fungal complex’ (Sneh & al. 1991) [Rhizoctonia DC.]. Genera such as Tulasnella J.Schröt—anamorph, Epulorhiza Moore—, Ceratobasidium D.P.Rogers—anamorph, Ceratorhiza Moore—, Thanatephorus Frank—anamorph, Monilliopsis Moore—, and Serendipita (Overw) P.Roberts, have been related with orchids (Taylor & al. 2003; Ortega-Larrocea & González, 2008). Mycorrhizal fungi are not usually fertile, so their classification is difficult and it is based on non-sexual characters, which allows their classification in morphotypes (Valadares & al. 2012, 2015). Thus to identify the mycorrhizal fungi, it is necessary to complement the morphological studies with molecular analyses (Valadares & al. 2012, 2015). In addition, Cruz & al. (2014) recorded the presence of cryptic species in Tulasnella, one of the most common genus forming mycorrhizas with terrestrial orchids.
Barranca del Cupatitzio Natural Reserve is located in the mexican state of Michoacán, between 19º25′ N and 19º26′19″ N, and 102º04′06″ W and 102º07′07″ W. The main vegetation of this zone includes pine forests, mixed forests of pine-oak, and a small area of cloud forest, as well as secondary vegetation. According with Bello-González & Madrigal-Sánchez (1996) and Zavala-Álvarez (2006), the pine forest is represented by species such as Pinus douglasiana Martínez, Pinus michoacana var. cornuta Martínez, Pinus lawsonii Roezl ex Gordon, Pinus leiophylla Schiede ex Schltdl. & Cham., Pinus pseudostrobus Brongn., Pinus pringlei Shaw, and Pinus oocarpa Schiede ex Schltdl., the three last ones being scarce; the mixed forest includes Quercus obtusata Bonpl., Quercus castanea Née, Quercus resinosa Liebm., Quercus candicans Née, Quercus magnoliifolia Née, Ceanothus caeruleus Lag., Coriaria ruscifolia L., Melampodium perfoliatum (Cav.) Kunth, Monnina schlechtendaliana D.Dietr., Phytolacca icosandra L., Salvia mexicana L., Verbesina oncophora B.L.Rob. & Seaton, Achimenes antirrhina (DC.) C.V.Morton, Alchemilla pringlei (Rydb.) Fedde, Commelina coelestis Willd., Crotalaria pumila Ortega, Cunila lythrifolia Benth., Drymaria villosa Schltdl. & Cham., Heterotheca inuloides Cass., Jaegeria hirta (Lag.) Less., Phaseolus acutifolius A.Gray, Ranunculus petiolaris Humb. & al. ex DC., Salvia lavanduloides Kunth, Spermacoce ocymoides Burm. f., Arbutus xalapensis Kunth, Bursera bipinnata (DC.) Engl., Lobelia laxiflora Kunth, Lupinus bilineatus Benth., Senecio angulifolius DC., Solanum lanceolatum Cav., Adiantum andicola Liebm., Asclepias glaucescens Kunth, Asclepias otarioides E.Fourn., Begonia gracilis Kunth, Drymaria villosa Schltdl. & Cham., Lopezia racemosa Cav., Muhlenbergia ciliata (Kunth) Trin., Muhlenbergia diversiglumis Trin., Pereilema crinitum J.Presl, Phaseolus coccineus L., Piqueria trinervia Cav., Plantago autralis Lam., Rhynchelytrum repens (Willd.) C.E.Hubb., Salvia elegans Vahl, Sisyrinchium cernuum (E.P. Bicknell) Kearney, and Sigesbeckia jorullensis Kunth; some representative species of the cloud forest and the secondary vegetation are Alnus jorullensis Kunth, Carpinus caroliniana Walter, Clethra mexicana DC., Ilex tolucana Hemsl., Fraxinus uhdei (Wenz.) Lingelsh., Hedyosmum mexicanum C.Cordem., Bocconia arborea S.Watson, Oreopanax salvinii Hemsl., Ternstroemia lineata DC., and Prunus capuli Cav. In Michoacán, a part of the forest ecosystems has been replaced by orchards of avocado, with a subsequent loss of the population of orchids.
In this context, the aims of our work were: (1) to analyze the variability of fungal species in terrestrial orchids of the genus Bletia in Mexico; (2) to know the systematic relationships among them; and (3) to elucidate if this variability is related to a wider distribution of these orchids in order to establish conservation strategies.
MATERIAL AND METHODSTOP
SamplingTOP
Three species of Bletia were selected: B. roezlii Rchb. f. and B. purpurata A.Rich. & Galeotti, which have a wide distribution in the temperate zones of Mexico, Guatemala, and Honduras, and B. punctata Lex., an endemic orchid from Mexico. The roots of 15–20 plants per species were collected at the flowering season of these species—from July to December—during five years—2010–2015—, allowing their correct identification. Three to five roots per plant were analyzed to obtain a total of 70 roots with fungal colonization from B. roezlii, 69 from B. purpurata, and 51 from B. punctata. The identification of the orchids was performed in the Morelia Orchidarium—Michoacán, Mexico (table 1).
Table 1. Identity of isolates, morphological and molecular identity of samples, GenBank accession number, host, reference sequences of identity and percentage (ID). [Sequenced samples are marked with an asterisk; M.A. Beltrán-Nambo collected the roots from the species of orchids and isolated the 39 strains].
| Fungal strain No. |
Host |
Morphological identification |
Molecular identity |
GenBank accession No. |
ID (%) |
Reference of the sequence |
| Subgroup A |
| 14A1 |
B. roezlii |
Tulasnella sp. 1 |
— |
— |
— |
— |
| 14D4 |
B. roezlii |
Tulasnella sp. 1 |
— |
— |
— |
— |
| 14D5* |
B. roezlii |
Tulasnella sp. 1 |
Tulasnella sp. |
MG008677 |
97 |
EF393627.1 |
| 19A1 |
B. purpurata |
Tulasnella sp. 1 |
— |
— |
— |
— |
| 19B1* |
B. purpurata |
Tulasnella sp. 1 |
Tulasnella sp. |
MG008679 |
97 |
EF393627.1 |
| 19C3 |
B. purpurata |
Tulasnella sp. 1 |
— |
— |
— |
— |
| 35A1 |
B. roezlii |
Tulasnella sp. 1 |
— |
— |
— |
— |
| Subgroup B |
| 14F2 |
B. roezlii |
Tulasnella sp. 2 |
— |
— |
— |
— |
| 46A1 |
B. roezlii |
Tulasnella sp. 2 |
— |
— |
— |
— |
| 47C1* |
B. punctata |
Tulasnella sp. 2 |
T. calospora |
MG008683 |
99 |
AY373286.1 |
| 48A2 |
B. purpurata |
Tulasnella sp. 2 |
— |
— |
— |
— |
| 48B1 |
B. purpurata |
Tulasnella sp. 2 |
— |
— |
— |
— |
| 49D1 |
B. roezlii |
Tulasnella sp. 2 |
— |
— |
— |
— |
| Subgroup C |
| 13A7 |
B. purpurata |
Tulasnella sp. 3 |
— |
— |
— |
— |
| 13B1 |
B. purpurata |
Tulasnella sp. 3 |
— |
— |
— |
— |
| 13C1* |
B. purpurata |
Tulasnella sp. 3 |
T. calospora |
MG008676 |
78 |
GU166407.1 |
| 13C2 |
B. purpurata |
Tulasnella sp. 3 |
— |
— |
— |
— |
| 16A2 |
B. purpurata |
Tulasnella sp. 3 |
— |
— |
— |
— |
| 16B4 |
B. purpurata |
Tulasnella sp. 3 |
— |
— |
— |
— |
| 27C1* |
B. roezlii |
Tulasnella sp. 3 |
T. calospora |
MG008680 |
96 |
GU166407.1 |
| 37A6* |
B. punctata |
Tulasnella sp. 3 |
T. calospora |
MG008681 |
98 |
JQ247558.1 |
| 37B2 |
B. punctata |
Tulasnella sp. 3 |
— |
— |
— |
— |
| 44D1 |
B. punctata |
Tulasnella sp. 3 |
— |
— |
— |
— |
| Subgroup D |
| 13A3 |
B. purpurata |
Tulasnella sp. 4 |
— |
— |
— |
— |
| 13B2* |
B. purpurata |
Tulasnella sp. 4 |
T. calospora |
MG008675 |
98 |
HQ889722.1 |
| 13B3 |
B. purpurata |
Tulasnella sp. 4 |
— |
— |
— |
— |
| 13SN |
B. purpurata |
Tulasnella sp. 4 |
— |
— |
— |
— |
| 16AST |
B. purpurata |
Tulasnella sp. 4 |
— |
— |
— |
— |
| 16B3 |
B. purpurata |
Tulasnella sp. 4 |
— |
— |
— |
— |
| 16B5* |
B. purpurata |
Tulasnella sp. 4 |
T. calospora |
MG008678 |
99 |
AB369439.1 |
| 18B1 |
B. roezlii |
Tulasnella sp. 4 |
— |
— |
— |
— |
| 18B3 |
B. roezlii |
Tulasnella sp. 4 |
— |
— |
— |
— |
| 23C1 |
B. punctata |
Tulasnella sp. 4 |
— |
— |
— |
— |
| 37A1 |
B. punctata |
Tulasnella sp. 4 |
— |
— |
— |
— |
| 37B3 |
B. punctata |
Tulasnella sp. 4 |
— |
— |
— |
— |
| 46AV |
B. roezlii |
Tulasnella sp. 4 |
— |
— |
— |
— |
| 46D |
B. roezlii |
Tulasnella sp. 4 |
— |
— |
— |
— |
| 46D1* |
B. roezlii |
Tulasnella sp. 4 |
T. calospora |
MG008682 |
98 |
FJ613176.1 |
| 50AST |
B. purpurata |
Tulasnella sp. 4 |
— |
— |
— |
— |
Isolation of mycorrhizal fungiTOP
The roots were washed and cross sections were made every 10 mm for long roots—≥ 12 cm—and every 6–8 mm for shorter roots—<
12 cm—. To select colonized segments, the samples were mounted on microscope slides after being fixed with Polyvinyl Alcohol Lactoglycerol—PVLG—. The surface of the colonized segments was disinfected immersing the roots in a 10% chlorine dilution, followed by an antibiotic solution—erythromycin 2% and gentamicin 1%—and rinses with sterile distillate water (Ortega-Larrocea 2008).
In vitro culturesTOP
Every disinfected root was transferred to a Petri dish with 1 ml of sterile distillate water. Velamen was removed from cortex, under laminar flow hood, with needle, and scalpel and pelotons were separated. Drops containing a solution of 10–15 pelotons were put in Petri dishes with a basic isolation medium for fungi—MAF—and a MAF sugar enriched medium—MAF-A—(Clements 1988; Mitchell 1989); Petri dishes were incubated under dark conditions at 27ºC until the hyphae emerging from pelotons were observed. Finally, the fragments of medium with the apexes of the hyphae were cut and transferred to a Papa-Dextrose Agar medium—PDA—with 50 mg/l of streptomycin and pH 6.8, the strains were maintained at 27ºC until the potential mycorrhizal cultures were obtained—they were morphologically verified by presence of septate hyphae, 90º ramifications with constriction and no sporulation—(Currah & al. 1987; Shan & al., 2002).
Morphological and statistical analysisTOP
The strains were incubated on PDA media for 30 days; during this period, the macroscopic and microscopic characterization of the morphology was performed. The cultures obtained from this study are store in the Laboratory of Genetics and Microbiology of the Faculty of Biology at the Universidad Michoacana de San Nicolás de Hidalgo.
The qualitative characteristics, such as surface color of the cultures, was determined after 15 days employing a Munsell Color Chart (Munsell 2000); the brightness, the texture, the odor, the growing shape, and the enzymatic capacity (Pereira & al. 2005) were analyzed. An enzymatic test with the tannic acid medium proposed by Davidson & al. (1938) and Zelmer (1994) was used to prove the presence of polyphenol oxidases. For each PDA isolate strain, three plates were inoculated with 1 mm3 of mycelia and incubated 5–15 days at 25ºC; cultures showing the change of color were considered as positives.
Quantitative characteristics such as the growth rates were evaluated by the technique of Currah & al. (1987), and the four-way radial growing increment of colonies was measured every 24–48 h during 2–8 weeks. The average values of growth rates were reported, based on three replicates per strain. The number of nuclei and the morphology of the hypha were determined as follows: media of 1 mm3 were removed from the PDA isolates and translated into Papa-Dextrose Broth medium—PDB—, the culture were maintained in agitation at 25ºC 15–20 days. To determine the number of nuclei, the fungal hyphae from PDB cultures were liquefied 1 min in 150 ml of distillated water, and aliquots of 1 ml were transferred to dialysis membranes mounted in a vacuum—minifall—. The membranes were fixed with 200 µl of a 2% formaldehyde solution for 20 min. The hyphae were stained using 200 µl of 4´,6´diamidino-2-phenylindole—DAPI—
5 µg/ml, for 20 min in darkness and washed with distillated water for 2 min. Previous to epifluorescence microscopic observations—200–400 nm UV wavelength—the samples were mounted on slides and fixed with 100 µl of glycerin 50% (Sneh & al. 1991). The sclerotia and the formation of monillioid cells were determined in 30 days-PDA cultures (Shan & al. 2002). Samples were mounted on slides after a trypan blue or acid fuchsine stain; the cell forms besides long and wide measurements using the Leica microscopy Z1000 with integrate camera, and then analyzed with an AMScope v. 3.7 program.
In order to determine significant differences between isolates, quantitative characteristics such as growth rates, monillioid cells, and hyphae dimensions were compared using ANOVA and Tukey tests with the program JMP v. 8. Strains were grouped throughout WARD agglomerative criterion based on Euclidian distances from qualitative and quantitative characteristics, cophenetic correlation was estimated in R Language v. 3.3 (R-Development Core Team 2008).
DNA extraction, amplification, and sequencingTOP
Since the strains in each group showed similar morphological characteristics and were extracted from the same root or plant, only some random strains from each previous formed group and from each orchid species were selected for this analysis. The DNA was isolated from fresh tissues previously grown in a PDB medium and vacuum pump washed, using a DNeasy Plant Mini Kit—Qiagen—.
The amplifications were performed using the universal primers for ITS1 and ITS4 (White & al. 1990) and following the protocol by Swarts & al. (2010), but with a Qiagen DNA-polymerase. Both forward and reverse sequencing was performed by Macrogen Korea Company.
Alignment of DNA sequence data and phylogenetic analysisTOP
The nine sequences obtained in this study were aligned with the most similar sequences available from GenBank—https://www.ncbi.nlm.nih.gov—in MUSCLE software (Edgar 2004), and manually improved with the program PhyDe® (Müller & al. 2005). Tulasnella danica Hauerslev was selected as the external group because its moderate divergence observed in the alignment and its location in the cladogram reported by Xing & al. (2013).
The program TNT V.1.5 was used for phylogenetic inferences (Goloboff & al. 2008). A Maximum Parsimony analysis was performed through an exhaustive search—Implicit Enumeration—and its statistical support was estimated with 10,000 bootstrap replications. In addition, employing the program MrBayes v. 3.2 (Ronquist & al. 2012), four Markov chains were run in parallel including 10,000,000 generations for each one, and using a GTR + G nucleotide substitution model that was estimated with the program jModeltest (Posada 2008).
RESULTSTOP
A total of 107 isolates were obtained from 190 processed roots of three species of Bletia. Those isolates that showed identical morphological characteristics of colonies and that were extracted from the same plant, were considered as one; eventually, 39 different strains were obtained, of which 19 came from B. purpurata, 13 from B. roezlii, and 7 from B. punctata (table 1). These 39 strains were classified into two groups and four subgroups according with the phenogram generated by the analysis WARD (fig. 1).
|
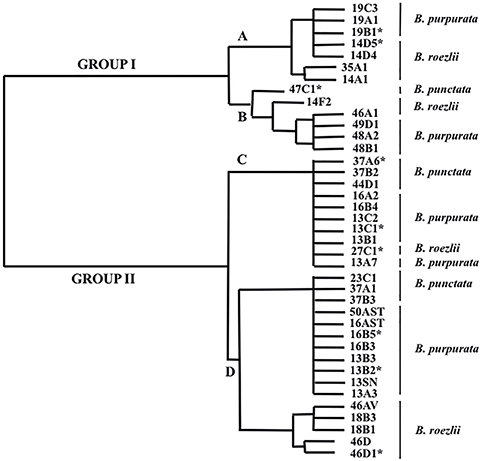 Fig. 1. Phenogram generated by the agglomerative algorithm WARD, using Euclidean distances derived from qualitative and quantitative characters of 39 isolated strains of mycorrhizal fungi—correlation coefficient 0.95—. [For each strain its corresponding species of orchid is shown; selected fungi for molecular analysis are marked with an asterisk.] Fig. 1. Phenogram generated by the agglomerative algorithm WARD, using Euclidean distances derived from qualitative and quantitative characters of 39 isolated strains of mycorrhizal fungi—correlation coefficient 0.95—. [For each strain its corresponding species of orchid is shown; selected fungi for molecular analysis are marked with an asterisk.]
|
|
Morphological descriptionTOP
The morphological characters of the isolated strains such as hyphae binucleate, branched in right angle and diameter of hyphae, septum near to branching point, frequent monilioid cells, polyphenol oxidase negative reaction, a slightly citric scent, etc., related them to the ‘Rhizoctonia-like fungal complex’ (fig. 2), specifically with the teleomorph genus Tulasnella, one of the most important mycobiont of the orchids (Oberwinkler & al. 2017). Nevertheless, it is important to note that—in the analyzed Mexican orchids—those mycorrhizal fungi included in subgroup A had lower hypha diameters compared to those registered in literature for Tulasnella (fig. 2d); and, those of the subgroup B showed monillioid cells with smaller dimensions contrasting with those described for the ‘Rhizoctonia-like fungal complex’ (fig. 2e).
|
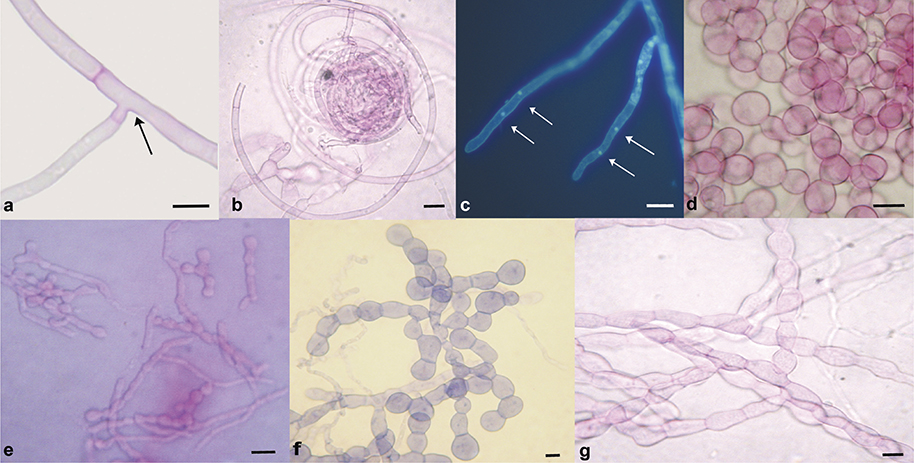 Fig. 2. Some morphological characters of the isolated strains: a, constriction at the branching point and septum near to branching point (strain 14D5); b, coils of hyphae (strain 19B1); c, DAPHI stained binucleate hyphae (arrows), epifluorescence microscopy (strain 47C1); d, monillioid cells (strain 19C3, subgroup A); e, monillioid cells (strain 49D1, subgroup B); f, monillioid cells (strain 27C1, subgroup C); g, monillioid cells, acid fuchsine staining (strain 23C1, subgroup D), acid fuchsine staining. Fig. 2. Some morphological characters of the isolated strains: a, constriction at the branching point and septum near to branching point (strain 14D5); b, coils of hyphae (strain 19B1); c, DAPHI stained binucleate hyphae (arrows), epifluorescence microscopy (strain 47C1); d, monillioid cells (strain 19C3, subgroup A); e, monillioid cells (strain 49D1, subgroup B); f, monillioid cells (strain 27C1, subgroup C); g, monillioid cells, acid fuchsine staining (strain 23C1, subgroup D), acid fuchsine staining.
|
|
The differences in qualitative characters such as the aspect of the colony and sclerotia (fig. 3) and the number of monillioid cells were observed in all the four subgroups. Some quantitative characteristics showed statistically significant differences between subgroups, like growth rates and the dimensions of the monillioid cells—P ≤ 0.01—(table 2). The strains of the group II exhibited significantly higher growth rates than those of the group I. The mycorrhizal fungi from the group II were isolated from the three studied species of orchids and from all the sampling sites; this can be considered as a viable option in the establishment of symbiosis in terrestrial orchids for conservation purposes.
Table 2. Quantitative morphological characterization of the strains obtained from roots of B. roezlli, B. punctata, and B. purpurata. [Different letters indicate significant differences between subgroups—P ≤ 0.05, Tukey—; n/d, no available data;
P, primary; S, secondary.]
| Subgroups |
Growth average rates (mm)n = 91, P ≤ 0.05 |
Monillioid cells (µm)n = 1074, P ≤ 0.05 |
Size of the hyphae (µm)n = 198 |
| Length (µm) |
Width (µm) |
Width (µm) |
Dist. septum P |
Dist. septum S |
Angle (degrees) |
| A |
0.9 ± 0.6 b |
10.7 ± 3 b |
9.1 ± 2.3 b |
3.5 ± 0.7 |
10.6 ± 5 |
2.6 ± 0.9 |
89.9 ± 12.2 |
| B |
0.7 ± 0.5 b |
5.3 ± 0.4 c |
4.5 ± 0.3 c |
3.1 ± 0.5 |
12.4 ± 5.8 |
3.1 ± 0.9 |
89.1 ± 5 |
| C |
2.5 ± 1.4 a |
13.9 ± 2 a |
11.7 ± 1.9 a |
3.7 ± 0.8 |
12.6 ± 7 |
3.1 ± 1.1 |
90.2 ± 9 |
| D |
2.2 ± 0.6 a |
13 ± 2 a |
8.4 ± 1.3 b |
3.6 ± 0.7 |
n/d |
n/d |
90 ± 2 |
|
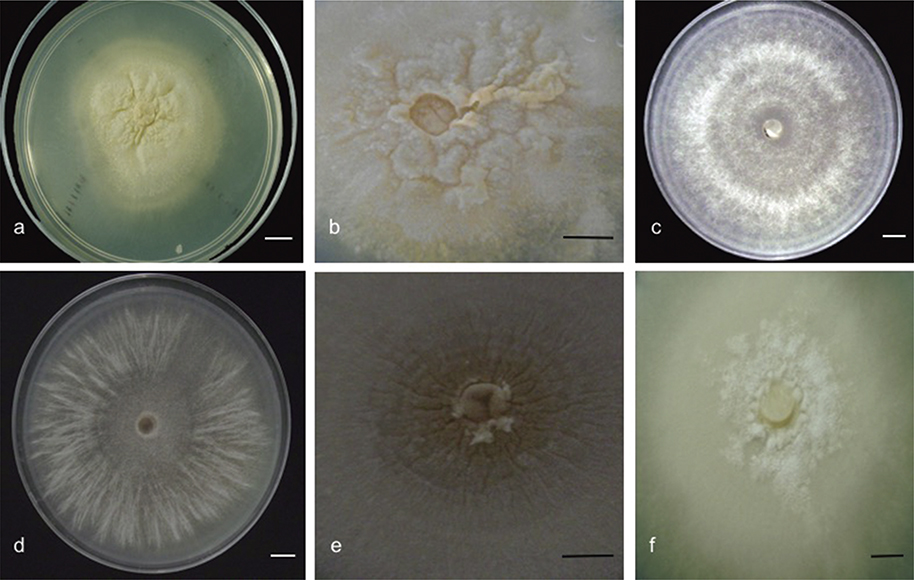 Fig. 3. Development of the different morphotypes obtained on the PDA culture: a, irregular and submerged growth of the subgroup A
(strain 19A1); b, waxy sclerotia and scabby appearance (strain 19B1); c, radial growth and mycelial ring formation, showing cottony colonies in subgroup B (strain 48B1); d, Cottony mycelium and strips-like formations in subgroup D (strain 16B5); e, submerged sclerotia, with sepia color and without scab formation in subgroup D (strain 16B3); f, uniform radial growth, colonies with waxy aspect, hyphae submerged in medium and sclerotia with granular appearance in subgroup C (strain 13C1). Fig. 3. Development of the different morphotypes obtained on the PDA culture: a, irregular and submerged growth of the subgroup A
(strain 19A1); b, waxy sclerotia and scabby appearance (strain 19B1); c, radial growth and mycelial ring formation, showing cottony colonies in subgroup B (strain 48B1); d, Cottony mycelium and strips-like formations in subgroup D (strain 16B5); e, submerged sclerotia, with sepia color and without scab formation in subgroup D (strain 16B3); f, uniform radial growth, colonies with waxy aspect, hyphae submerged in medium and sclerotia with granular appearance in subgroup C (strain 13C1).
|
|
Molecular identificationTOP
The phylogenetic analysis of the nine obtained sequences (fig. 4) in this study and the six closest Gen Bank sequences identified as three species of the genus Tulasnella—T. deliquescens (Juel) Juel, T. calospora (Boud.) Juel, and T. bifrons Bourdot & Galzin—and T. danica as the external group, gave rise to a phylogenetic tree with three clades. The clade I was integrated by sequences of the subgroups A and B of the morphological analysis and they were obtained from specimens from the three orchid species (table 1).They were grouped in two subclades, as they were in the morphological analysis (fig. 1), showing more than a 90% of identity when they were compared with the sequences of NCBI database, named here as Tulasnella sp. 1 and Tulasnella sp. 2. The sequences of the strains of subgroup C are grouped in the clade II, next the sequences identified as T. calospora and T. deliquescens. The sequences of subgroup D were grouped in the clade III, all of them with the sequences identified as T. calospora in a subclade with T. bifrons as sister group.
|
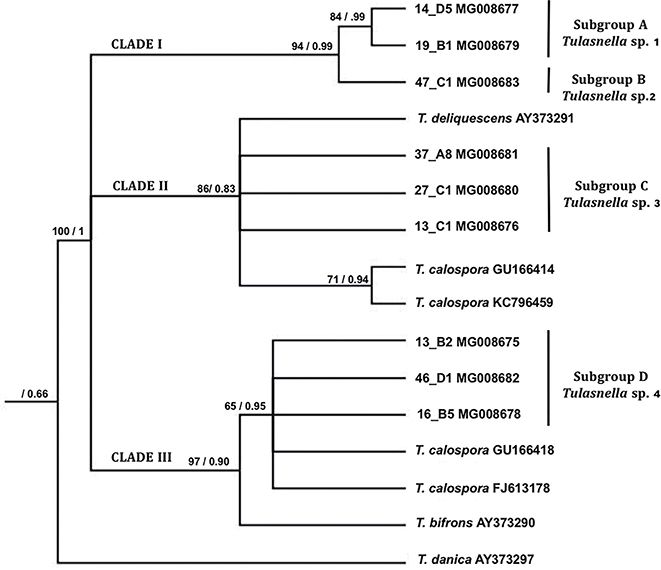 Fig. 4. Phylogenetic inference generated by Maximum Parsimony—Consistency Index 0.764, Retention Index 0.783—, ratified with Bayesian analysis. [The nodes with statistical support show bootstrap values/a posteriori probabilities. The relationships between the sequences generated in this study and the subgroups obtained from the morphological analysis, as well as the NCBI-referenced sequences are indicated.] Fig. 4. Phylogenetic inference generated by Maximum Parsimony—Consistency Index 0.764, Retention Index 0.783—, ratified with Bayesian analysis. [The nodes with statistical support show bootstrap values/a posteriori probabilities. The relationships between the sequences generated in this study and the subgroups obtained from the morphological analysis, as well as the NCBI-referenced sequences are indicated.]
|
|
TaxonomyTOP
Tulasnella sp. 1. Subgroup A.
Isolates waxy flat to plush, irregular and submerged growth, sometime with slightly cottony appearance and irregular growth through the subsequent cultures, with waxy glabrous sclerotia and scabby appearance (fig. 3a, b). Hyphae hyaline, thin-walled, diameter 3.5–4.2 µm. Growth rate 0.9 mm/day, similar to subgroup B, but significantly different to subgroups C and D included in group 2—P ≤ 0.01—, reaching their maximum development in Petri dishes at 40 days or more.
Monillioid cells with intermediate sizes—10.7—9.1 µm—compared with Tulasnella sp. 3 and Tulasnella sp. 4 (P ≤ 0.01), 3–5 cells in chain (fig. 2d, table 2).
Tulasnella sp. 2. Subgroup B.
Strains with radial growth and mycelial ring formation with cottony aspect on medium, white to pale yellow—8/2 to 8/3 5Y—(fig. 3c). Hyphae hyaline thin-walled, diameter 3.1 µm. Growth rate 0.9 mm/day and maximum development in Petri dish after 40 days (table 2). Diameter of monillioid cells 5.3 × 4.5 µm, smaller than reported to this genus—10–25 × 25–40 µm (Currah & al. 1997)—, 5–10 cells in chain (fig. 2e).
Tulasnella sp. 3. Subgroup C.
Colonies with waxy appearance, with sclerotia scattered over colony occasionally conferring a granular appearance (fig. 3f); colony and sclerotia white to pale yellow—8/2 5Y—. Growth rate 2.5 mm/day, significantly higher than those of Tulasnella sp. 1 and Tulasnella sp. 2—P ≤ 0.01—; maximum development in Petri dish is reached in 15–20 days. Hyphae hyaline with diameter 3.6–4.4 µm. Monillioid cells spherical, with diameter 14 µm, with 3–7 cells in chain, significantly different to all other subgroups—P ≤ 0.01—(fig. 2f, table 2). This morphological characters related this species to T. calospora.
Tulasnella sp. 4. Subgroup D.
Colonies with radial growth and mycelial ring aspect (fig. 3d); submerged sclerotia, brown—7/4 2.5 Y to 8/2 5Y—and without scab formation (fig. 3e). Growth rate 2.2 mm/day growth, values—P ≤ 0.01—, significantly higher than those of Tulasnella sp. 1 and Tulasnella sp. 2; maximum development in Petri dish in 15–20 days. Hyphae dimensions were similar to those of Tulasnella sp. 3—3.6–4.4 µm—, but with oval monillioid cells, 3–5 in chain (fig. 2g, table 2). This morphological characters related this species to T. calospora.
DISCUSSIONTOP
Our results corroborate those previously obtained for other genera of orchids such as Orchis L., Platanthera Rich., and Tipularia Nutt. (Dearnaley & al. 2012; Pandey & al. 2013), in which the association with different mycorrhizal fungi in the same root was more common among photosynthetic orchids than among mycoheterotrophic orchids, because two or more morphotypes from the same root were isolated from B. roezlii and B. purpurata. This fact may confer to plants an advantage on obtainment of nutrients and a greater capability to survive in the environment (Mageto & al. 2014). Seven morphotypes included in three clades were obtained from the endemic species B. punctata, whereas for the other two orchid species included in this study, more than 13 morphotypes were isolated and grouped into the four subclades; this agrees with some observations that pointed out that orchids with a wide distribution range have associations with generalist fungi, conversely to orchids with a restricted distribution, which may have specific relations with fungi (Mageto & al. 2014; McCormick & Jacquemyn 2014).
Morphotypes of clade II displayed relationship with the reference sequences identified as T. calospora, T. deliquescens, and T. bifrons; the two last ones have been found associated to other adult photosynthetic terrestrial orchids from Canada and North America such as Tipularia discolor (Pursh) Nutt. and Goodyera pubescens (Willd.) R.Br., mainly in forests of pine-oaks (Rasmussen & Rasmussen 2007), which is comparable to the sampling zones of this work; McCormick & Jacquemyn (2014) pointed out that mycorrhizal associations in a large number of orchid species have corroborated that fungal symbionts can exhibit a wide distribution range and they are able to adapt to different habitats.
The molecular and morphological analyses allowed us to identify the selected strains as different clades of Tulasnella, a genus included in the ‘Rhizoctonia-like fungal complex’ (Valadares & al. 2012; Nogueira & al. 2014; Suárez & Kottke 2016). Rhizoctonia has been described around the world in a wide range of photosynthetic orchids species such as Epidendrum secundum Jacq., Acianthera lamia (Luer) Pridgeon & M.W.Chase, Polystachya concreta (Jacq.) Garay & H.R.Sweet, among others (Zettler & al. 2004; Pereira & al. 2005; Nogueira & al., 2014), and it has also been documented in the Mexican photosynthetic orchids B. urbana Dressler and B. campanulata Lex. (Ortega-Larrocea & Rangel-Villafranco 2007).
All fungal morphotypes isolated in this work were assigned to the genus Tulasnella and grouped in separate clades related to different species, showing that plants of Bletia from the same population can be associated with different species of fungi at the same time, at least during the adult stage, which has also been reported for other orchid genera such as Dendrobium Sw., Orchis, and Liparis Rich. (Cruz & al. 2014; Suárez & Kottke 2016). This versatility of fungal partner can contribute to the abundance and distribution of the population of the orchid, as previously reported, and it depends on biotic and abiotic factors, including the availability of suitable mycorrhizal fungi contributing to the health of the plants (Beltrán-Nambo & al. 2012; Jacquemyn & al. 2012; Xing & al. 2013; McCormick & Jacquemyn 2014; Kumar & al. 2017).
According with Pereira & al. (2014), this study corroborated that T. calospora is a species complex. The sequences of the clades II and III were grouped with different sequences identified as T. calospora, so we cannot assign this name to either of two clades until we confirm in which of them the type is located. Other works reported the presence of cryptic species in Tulasnella (Cruz & al. 2014, 2016). In order to elucidate the diversity and variability of the species of mycorrhizal fungi, additional studies using molecular and morphological approaches are required for more mycorrhizal fungi in orchids. In this way, new species could be described and the diversity of mycorrhizal fungi could be used for conservation purposes.
ACKNOWLEDGEMENTSTOP
This work was financed by CECTI project No. 05. Authors are thankful to Jesús Cruz, and PhD Rafael Salgado Garciglia for the identification of the orchids during the development of the project Orquídas del Parque Nacional Barranca del Cupatitzio. To M.C. Aarón Giovanni Munguía Rodríguez for helping to improve the translation of this article to English, and to PhD. Pilar Ortega Larrocea for her transfer of knowledge that made this work possible.
REFERENCESTOP
| ○ |
Bello-González M.A & Madrigal-Sánchez X. 1996. Estudio florístico del campo experimental “Barranca del Cupatitzio”, Uruapan, Michoacán. INIFAP [folleto científico n.º 2], Uruapan.
|
| ○ |
Beltrán-Nambo M.A., Ortega-Larrocea M.P., Salgado-Garciglia R., Otero-Ospina J.T., Martínez-Trujillo M. & Carreón-Abud Y.
2012. Distribution and abundance of terrestrial orchids of the genus Bletia in sites with different degrees of disturbance, in the Cupatitzio Natural Reserve, Mexico. International Journal of Biodiversity and Conservation 4 (8): 316–325.
|
| ○ |
Clements M.A. 1988. Orchid mycorrhizal associations. Lindleyana 3: 73–86.
|
| ○ |
Cruz D., Suárez J.P. & Piepenbring M. 2014. Cryptic species revealed by molecular phylogenetic analysis of sequences obtained from basidiomata of Tulasnella. Mycologia 106 (4): 708–722. https://doi.org/10.3852/12-386 |
| ○ |
Cruz D., Suárez J.P., Kottke I. & Piepenbring M. 2016. Morphological revision of Tulasnellaceae, with two new species of Tulasnella and new records of Tulasnella spp. for Ecuador. Nova Hedwigia 102 (3–4): 279–338. https://doi.org/10.1127/nova_hedwigia/2015/0304 |
| ○ |
Currah R.S., Sigler L. & Hambleton S. 1987. New records and new taxa of fungi from the mycorrhizae of terrestrial orchids of Alberta. Canadian Jounal of Botany 65: 2473–2482. https://doi.org/10.1139/b87-336 |
| ○ |
Currah R.S., Zelmer C.D., Hambleton S. & Richardson K.A. 1997. Fungi from orchid mycorrhizas. In Arditty J. & Pridgeon A. (eds.), Orchid Biology, Reviews and Perspectives VII. Kluwer Academic Publishers, Dordrecht. https://doi.org/10.1007/978-94-017-2498-2_4 |
| ○ |
Davidson R.W., Cambell W.A. & Blaisedel D.J. 1938. Differentiation of wood-decaying fungi by their reactions on gallic or tannic acid medium. Journal Agriculture Research 57: 683–695.
|
| ○ |
Dearnaley J.D., Martos F. & Selosse M.A. 2012. Orchid mycorrhizas: Molecular ecology, evolution and conservation aspects.
In Hock B. (ed.), The Mycota IX: 207–230. Springer, Berlin.
|
| ○ |
Edgar R.C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. Bioinformatics 5 (113): 1–19.
|
| ○ |
Goloboff P.A., Farris J.S. & Nixon K.C. 2008. TNT, a free program for phylogenetic analysis. Cladistics 24: 774–786. https://doi.org/10.1111/j.1096-0031.2008.00217.x |
| ○ |
Govaerts R., Bernet P., Kratochvil K., Gerlach G., Carr G., Alrich P., Pridgeon A.M., Pfahl J., Campacci M.A., Baptista D.H.,
Tigges H., Shaw J., Cribb P., George A., Kreuz K. & Wood J. 2016. World Checklist of Orchidaceae Family. Website: http://apps.kew.org/wcsp/ [accessed XI-2016].
|
| ○ |
Jacquemyn H., Brys R., Lievens B. & Wiegand T. 2012. Spatial variation in below-ground seed germination and divergent mycorrhizal associations correlate with spatial segregation of three co-occurring orchid species. Journal of Ecology 100 (6): 1328–1337. https://doi.org/10.1111/j.1365-2745.2012.01998.x |
| ○ |
Kumar P.M., Pagano M. & Donovan O. 2017. Biotechnological advances of beneficial fungi for plants. Mycosphere 8 (3): 445–455. https://doi.org/10.5943/mycosphere/8/3/6 |
| ○ |
Mageto O.E., Kamweya M.A., Ochora M.J. & Maobe S.N. 2014. Effects of symbiotic fungi on distribution and abundance of the epiphyitic orchid Polystachya fusiformis (Thou.) Lindl. in the manga range ecosystem, Kisii, Kenya. Journal of Applied Phytotechnology in Environmental Sanitation 3 (2): 45–54.
|
| ○ |
McCormick M.K. & Jacquemyn H. 2014. What constrains the distribution of orchid populations? New Phytologist 202 (2): 392–400. https://doi.org/10.1111/nph.12639 |
| ○ |
Mitchell R. 1989. Growing hardy orchids from seed at Kew. The Plantsman 3 (2): 152–169.
|
| ○ |
Müller J., Muller K., Neinhuis C. & Quandt D. 2005. PhyDE: Phylogenetic Data Editor. Website: www.phyde.de [accessed 16-XII-2016]. |
| ○ |
Munsell. 2000. Soil Color Charts. GretagMacbeth. 617 Life Britain Road, New Windsor.
|
| ○ |
Nogueira R.E., van den Berg C., Pereira O.L. & Kasuya M.M. 2014. Isolation and molecular characterization of Rhizoctonia-life fungi associated with orchid roots in the Quadrilátero Ferrífero and Zona da Mata regions of the state of Minas Gerais,
Brazil. Acta Botánica Brasilica 28 (2): 298–300. https://doi.org/10.1590/S0102-33062014000200017 |
| ○ |
Oberwinkler F., Cruz D. & Suárez J.P. 2017. IBiogeogrphy and Ecology of Tullasnelaceae. In Caldwell M.M., Canadell J.G., Díaz S., Heldmaier G., Jackson R.B., Levia D.F., Schulze E.-D., Sommer U. & Wardle D.A. (eds.),
Ecological Studies 230: 237–271. Springer, Berlin.
|
| ○ |
Ortega-Larrocea M.P. 2008. Propagación simbiótica de orquídeas terrestres con fines de restauración edafológica. In Álvarez-Sánchez J. & Monroy-Ata A. (eds.), Técnicas de estudio de las asociaciones micorrízicas y sus implicaciones en la restauración: 85–96. Facultad de Ciencias UNAM, Ciudad de México.
|
| ○ |
Ortega-Larrocea M.P. & Rangel-Villafranco M. 2007. Fungus assisted reintroduction and lon-term survival of two Mexican terrestrial orchids in the natural habitat. Lankesteriana 7 (1–2): 317–321.
|
| ○ |
Ortega-Larrocea M.P. & González D. 2008. Los hongos asociados a las orquídeas terrestres en la restauración. In Heredia A.G. (ed.), Tópicos sobre diversidad, ecología y usos de los hongos microscópicos en Iberoamérica: 305. Instituto de Ecología A.C., Veracruz.
|
| ○ |
Otero J.T., Mosquera A.T. & Flanagan N.S. 2013. Tropical orchid mycorrizae: potential in orchid conservation, commercialization and beyond. Lankesteriana 13 (1–2): 57–63.
|
| ○ |
Pandey M., Sharma J., Taylor D.L. & Yadon V.L. 2013. A narrowly endemic photosynthetic orchid is non-specific in its mycorrhizal associations. Molecular Ecology 22 (8): 2341–2354. https://doi.org/10.1111/mec.12249 |
| ○ |
Pereira O.L., Megumi K.M., Chaer B.A. & Fernandes de Araújo A.E. 2005. Morphological and molecular characterization of mycorrhizal fungi isolated from Neotropical orchids in Brazil. Canadian Journal of Botany 83: 54–65. https://doi.org/10.1139/b04-151 |
| ○ |
Pereira M.C., Coelho I.S., Valadares R.B.S., Oliveira S.F., Bocayuva M., Pereira O.L., Araujo E.F. & Kasuya M.K. 2014. Morphological and molecular characterization of Tulasnella spp. fungi isolated from the roots of Epidendrum secundum, a widespread orchid. Symbiosis 62: 111–121. https://doi.org/10.1007/s13199-014-0276-0 |
| ○ |
Posada D. 2008. jModelTest: Phylogenetic Model Averaging. Molecular Biology and Evolution 25 (7): 1253–1256. https://doi.org/10.1093/molbev/msn083 |
| ○ |
R-Development Core Team. 2008. R: a language and environment for statistical computing. R Fundationfor Statistical Compiuting, Vienna. Website: http://www.R-proyect.org [accessed 5-IX-2016].
|
| ○ |
Rasmussen H.N. & Rasmussen F.N. 2007. Trophic relationships in orchid mycorrhiza-diversity and implications for conservation.
Lankesteriana 7 (1–2): 334–341.
|
| ○ |
Ronquist F., Teslenko M., Mark P.V., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A. & Huelsenbeck J.P.
2012. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Systematic Biology 61 (3): 539–542. https://doi.org/10.1093/sysbio/sys029 |
| ○ |
Schatz B., Geoffroy A., Dainat B., Bessière J., Buatois B., Hossaert-McKey M. & Selosse M. 2010. A case study of modified interactions with symbionts in a hybrid mediterranean orchid. American Journal of Botany 97 (8): 1278–1288. https://doi.org/10.3732/ajb.0900303 |
| ○ |
SEMARNAT. 2010. Norma Oficial Mexicana NOM-059-ECOL-2001: Protección ambiental-Especies de flora y fauna silvestres de México-Categorías de riesgo y especificaciones para su inclusión, exclusión. Lista de especies en riesgo. Diario Oficial de la Federación [6-III-2010], México.
|
| ○ |
Shan X.C., Liew E.C., Weatherhead M.A. & Hodgkiss J.J. 2002. Characterization and taxonomic placement or Rhizoctonia-like endophytes from orchid roots. Mycologia 94 (2), 230–239. https://doi.org/10.1080/15572536.2003.11833228 |
| ○ |
Sneh B., Burpee L. & Ogoshi A. 1991. Identification of Rhizoctonia species. American Phytopathological Society Press, Minnesota.
|
| ○ |
Sosa V. 1992. Neotificación de tres especies del género Bletia (Orchidaceae). Acta Botánica Mexicana 18: 71–79. https://doi.org/10.21829/abm18.1992.644 |
| ○ |
Suárez J.P. & Kottke I. 2016. Main fungal partners and different levels of specificity of orchid mycorrhizae in the tropical mountain forests of Ecuador. Lankesteriana 16 (2): 299–305. https://doi.org/10.15517/lank.v16i2.26014 |
| ○ |
Swarts N.D., Sinclair E.A., Francis A. & Dixon K.W. 2010. Ecological specialization in mycorrhizal symbiosis leads to rarity in an endangered orchid. Molecular Ecology 19: 3226–3242. https://doi.org/10.1111/j.1365-294X.2010.04736.x |
| ○ |
Taylor D.L., Bruns T.D., Szaro T.M. & Hodges S.A. 2003. Divergence in mycorrhizal specialization within Hexalectris spicata (Orchidaceae), a nonphotosynthetic desert orchid. American Journal of Botany 90: 1168–1179. https://doi.org/10.3732/ajb.90.8.1168 |
| ○ |
Valadares R.B.S., Pereira M.C., Otero J.T. & Cardoso E.J.B.N. 2012. Orchid mycorrhiza diversity in Coppensia doniana, a widespread Oncidiinae from campos de Jordao-SP Brazil. Biotropica 44: 114–122. https://doi.org/10.1111/j.1744-7429.2011.00769.x |
| ○ |
Valadares R.B.S., Otero J.T., Pereira M.C. & Cardoso E.J.B.N. 2015. The epiphytic orchid Ionopsis utricularioides and Psygmorchis pusilla associate with different Ceratobasidium lineajes at Valle de Cauca, Colombia. Acta Botanica Brasilica 29 (1): 40–44. https://doi.org/10.1590/0102-33062014abb3397 |
| ○ |
White T.J., Burns T., Lee S. & Taylor J. 1990. Amplification and direct secuencing of fungal ribosomal RNA genes for phylogenetics.
In Innis M.A., Gelfand D.H., Sninsky J.J. & White T.J. (eds.), PCR Protocols: A Guide to Methods and Applications: 315–322. Academic Press, New York.
|
| ○ |
Whitman M. &. Ackerman J.D. 2015. Terrestrial orchids in a tropical forest: best sites for abundance differ from those for reproduction. Ecology 96 (3): 693–704. https://doi.org/10.1890/14-0104.1 |
| ○ |
Xing X., Ma X., Deng Z., Chen J., Wu F. & Guo S. 2013. Specificity and preference of mycorrhizal associations in two species of the genus Dendrobium (Orchidaceae). Mycorrhiza 23: 317–324. https://doi.org/10.1007/s00572-012-0473-8 |
| ○ |
Zavala-Álvarez C. 2006. Pteridoflora del Parque Nacional Barranca del Cupatitzio, Uruapan, Michoacán, México. Dissertation,
Universidad Michoacana de San Nicolás de Hidalgo, Mexico.
|
| ○ |
Zelmer C.D. 1994. Interactions between northern terrestrial orchids and fungi in nature. PhD. Dissertation, University of Alberta, Canada.
|
| ○ |
Zettler L.W., Sharma J. & Rasmussen F. 2004. Mycorrhizal diversity. In Dixon K., Cribb P., Kell S. & Barret R. (eds), Orchid Conservation: 185–203. Natural History Publications, Kota Kinabalu.
|
Fig. 1. Phenogram generated by the agglomerative algorithm WARD, using Euclidean distances derived from qualitative and quantitative characters of 39 isolated strains of mycorrhizal fungi—correlation coefficient 0.95—. [For each strain its corresponding species of orchid is shown; selected fungi for molecular analysis are marked with an asterisk.]